Contact
Provider(s):
You must log in in order to see contact information. If you don't have an account, you can ask for one on this form.
Rapid detection of SARS-Cov-2 and E. coli (ESBL) via the utilisation of a portable SHERLOCK/DETECTR approach.
In STAMINA project, we have developed novel genetic markers allowing the fast detection of SARS-CoV-2 and Enterobacterales in human and environmental samples in a CRISPR-Cas based Dx assay (SHERLOCK/DETECTR approach). This approach allows the rapid detection of these pathogens while in most cases shows higher sensitivity compared to the standard RT-PCR.
- SARS-CoV-2 is a virus of the species severe acute respiratory syndrome–related coronavirus (SARSr-CoV). Epidemiological studies estimate that each infection results in an average of 2.39 to 3.44 new ones when no members of the community are immune, and no preventive measures are taken. The virus primarily spreads between people through close contact and via aerosols and respiratory droplets that are exhaled when talking, breathing, or otherwise exhaling, as well as those produced from coughs or sneezes.
- Enterobacterales are a large order of different types of bacteria (germs) that commonly cause infections both in healthcare settings and in communities. Some E. coli strains can produce enzymes called extended-spectrum beta-lactamases (ESBLs). ESBL enzymes break down and destroy some commonly used antibiotics, including penicillins and cephalosporins, and make these drugs ineffective for treating infections. This resistance means that there are fewer antibiotic options available to treat ESBL-producing Enterobacterales infections. In many cases, even common infections, such as urinary tract infections, caused by ESBL-producing germs require more complex treatments.
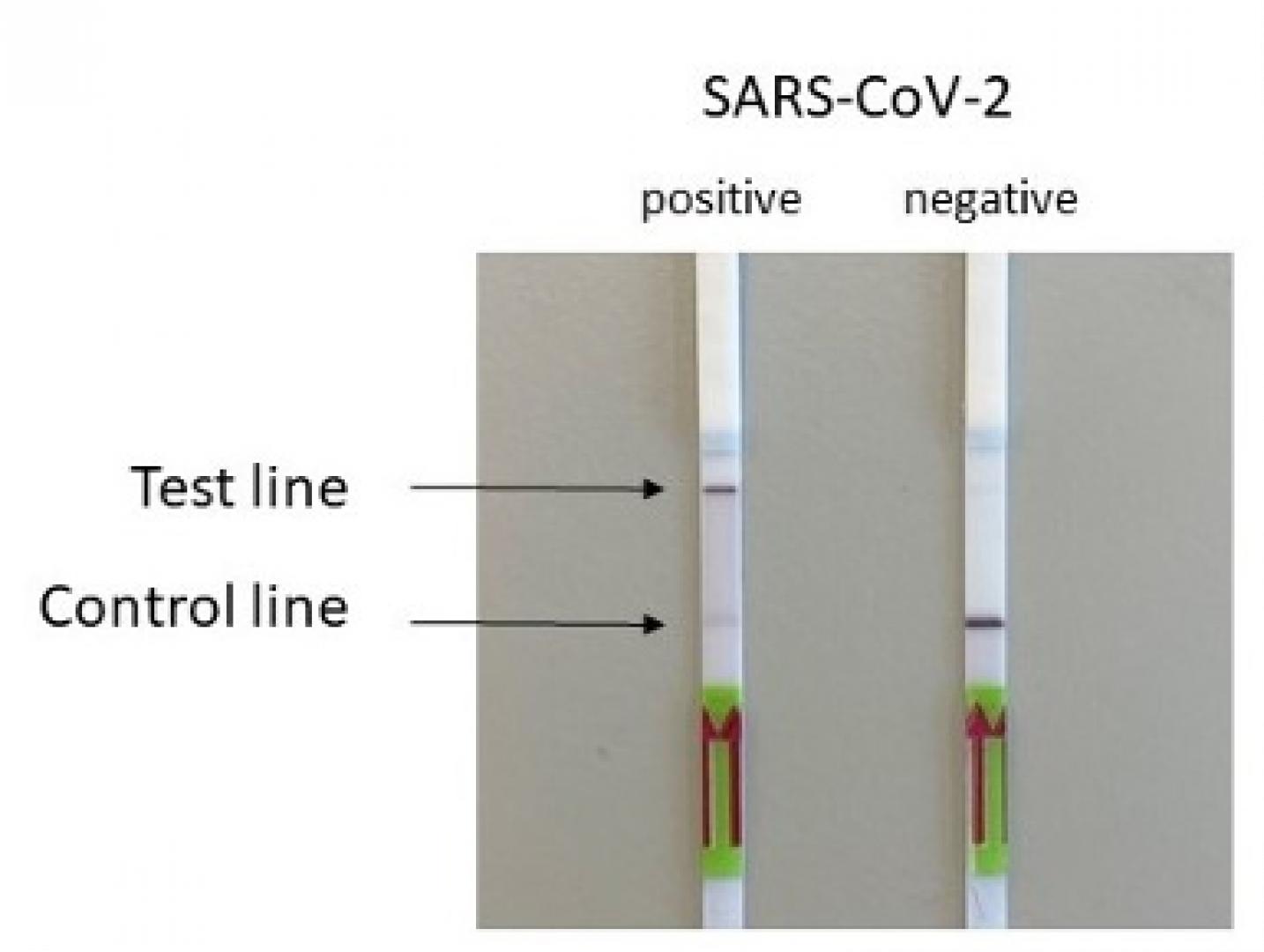
The HybridDetect Dipstick (illustration), resembles the rapid self-tests currently available, in terms of size.
Supported Use Cases
SHERLOCK/DETECTR based early warning and spread monitoring for viral and bacterial pathogens
SHERLOCK/DETECTR based rapid diagnostic for viral and bacterial pathogens
Addressed hazards
Innovation stage
Readiness
Crisis Cycle Phase
Crisis size
Illustrations
 |
Portfolio of Solutions web site has been initially developed in the scope of DRIVER+ project. Today, the service is managed by AIT Austrian Institute of Technology GmbH., for the benefit of the European Management. PoS is endorsed and supported by the Disaster Competence Network Austria (DCNA) as well as by the STAMINA and TeamAware H2020 projects. |